这篇文章主要介绍“GWAS怎么去掉亲缘关系近的个体”的相关知识,小编通过实际案例向大家展示操作过程,操作方法简单快捷,实用性强,希望这篇“GWAS怎么去掉亲缘关系近的个体”文章能帮助大家解决问题。
plink --bfile HapMap_3_r3_10 --genome --min 0.2 --out pihat_min0.2说明文档:
--genome invokes an IBS/IBD computation, and then writes a report with the following fields to plink.genome:FID1 Family ID for first sampleIID1 Individual ID for first sampleFID2 Family ID for second sampleIID2 Individual ID for second sampleRT Relationship type inferred from .fam/.ped fileEZ IBD sharing expected value, based on just .fam/.ped relationshipZ0 P(IBD=0)Z1 P(IBD=1)Z2 P(IBD=2)PI_HAT Proportion IBD, i.e. P(IBD=2) + 0.5*P(IBD=1)PHE Pairwise phenotypic code (1, 0, -1 = AA, AU, and UU pairs, respectively)DST IBS distance, i.e. (IBS2 + 0.5*IBS1) / (IBS0 + IBS1 + IBS2)PPC IBS binomial testRATIO HETHET : IBS0 SNP ratio (expected value 2)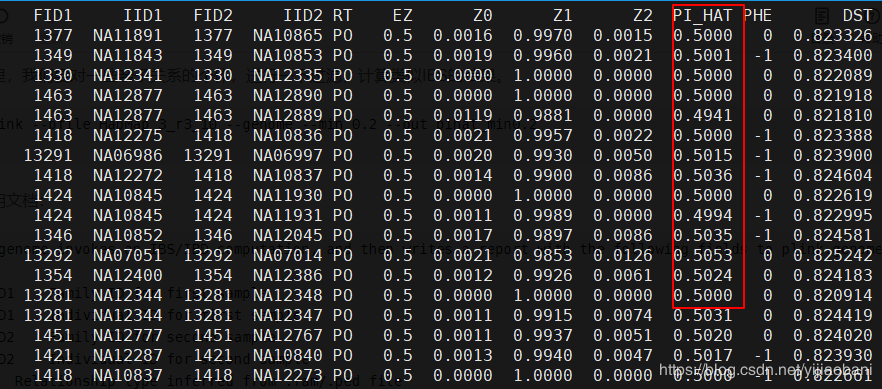
awk '{if($8>0.9) print $0}' pihat_min0.2.genome > zoom_pihat.genome过滤出91个组合: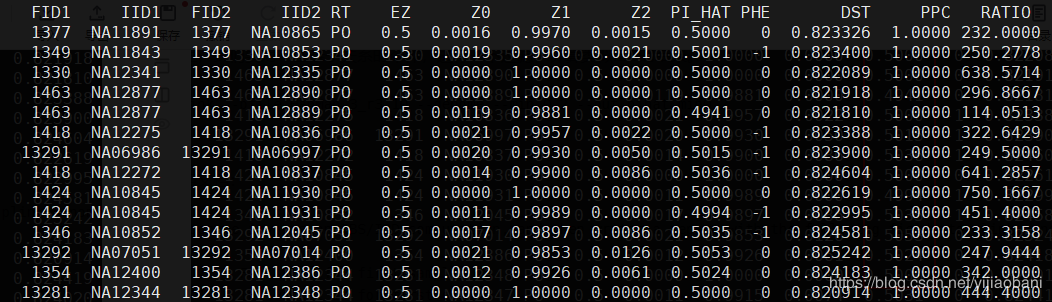
R代码
pdf("relatedness.pdf")relatedness = read.table("pihat_min0.2.genome", header=T)par(pch=16, cex=1)with(relatedness,plot(Z0,Z1, xlim=c(0,1), ylim=c(0,1), type="n"))with(subset(relatedness,RT=="PO") , points(Z0,Z1,col=4))with(subset(relatedness,RT=="UN") , points(Z0,Z1,col=3))legend(1,1, xjust=1, yjust=1, legend=levels(relatedness$RT), pch=16, col=c(4,3))pdf("zoom_relatedness.pdf")relatedness_zoom = read.table("zoom_pihat.genome", header=T)par(pch=16, cex=1)with(relatedness_zoom,plot(Z0,Z1, xlim=c(0,0.02), ylim=c(0.98,1), type="n"))with(subset(relatedness_zoom,RT=="PO") , points(Z0,Z1,col=4))with(subset(relatedness_zoom,RT=="UN") , points(Z0,Z1,col=3))legend(0.02,1, xjust=1, yjust=1, legend=levels(relatedness$RT), pch=16, col=c(4,3))pdf("hist_relatedness.pdf")relatedness = read.table("pihat_min0.2.genome", header=T)hist(relatedness[,10],main="Histogram relatedness", xlab= "Pihat")dev.off()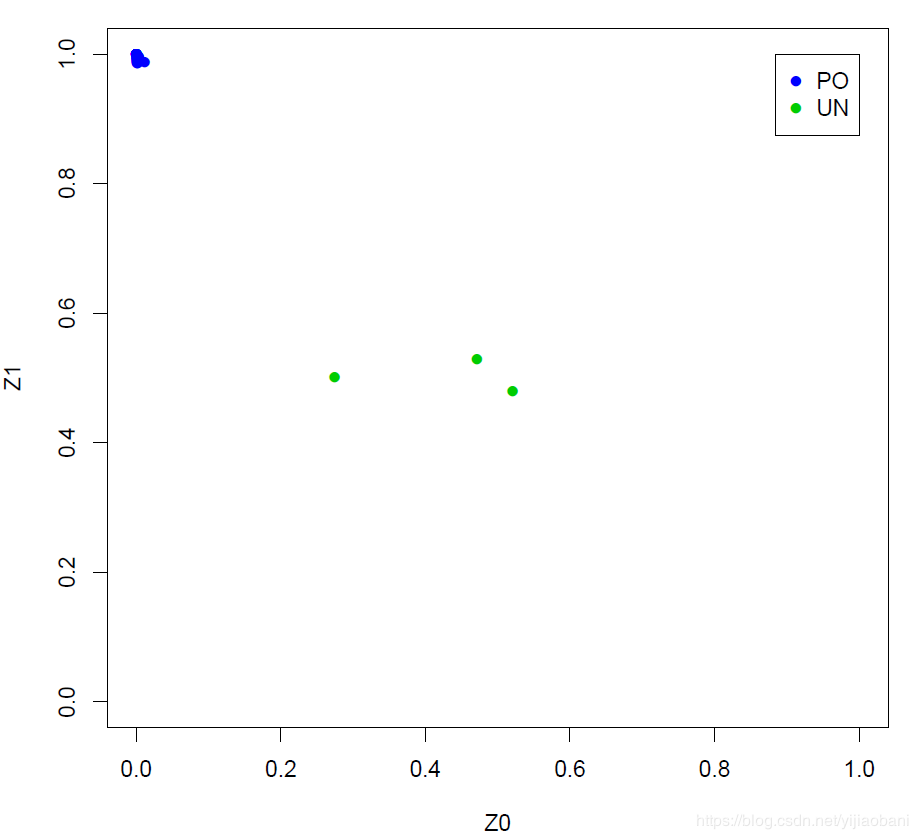 这里的PO为:亲子关系 这里的UN为:非亲缘关系
这里的PO为:亲子关系 这里的UN为:非亲缘关系
plink --bfile HapMap_3_r3_10 --filter-founders --make-bed --out HapMap_3_r3_11 日志:
PLINK v1.90b6.5 64-bit (13 Sep 2018) www.cog-genomics.org/plink/1.9/(C) 2005-2018 Shaun Purcell, Christopher Chang GNU General Public License v3Logging to HapMap_3_r3_11.log.Options in effect: --bfile HapMap_3_r3_10 --filter-founders --make-bed --out HapMap_3_r3_11515185 MB RAM detected; reserving 257592 MB for main workspace.1073743 variants loaded from .bim file.161 people (77 males, 84 females) loaded from .fam.110 phenotype values loaded from .fam.51 people removed due to founder status (--filter-founders).Using 1 thread (no multithreaded calculations invoked).Before main variant filters, 110 founders and 0 nonfounders present.Calculating allele frequencies... done.Total genotyping rate in remaining samples is 0.998016.1073743 variants and 110 people pass filters and QC.Among remaining phenotypes, 55 are cases and 55 are controls.--make-bed to HapMap_3_r3_11.bed + HapMap_3_r3_11.bim + HapMap_3_r3_11.fam ...done. 可以看出,51个个体被移除。
HapMap_3_r3_11.bed HapMap_3_r3_11.bim HapMap_3_r3_11.fam HapMap_3_r3_11.log这里讲亲子关系的个体移除,不是必须要的,比如我们分析的群体里面有亲子关系的个体,想要进行分析,不需要做这一步的筛选。
关于“GWAS怎么去掉亲缘关系近的个体”的内容就介绍到这里了,感谢大家的阅读。如果想了解更多行业相关的知识,可以关注亿速云行业资讯频道,小编每天都会为大家更新不同的知识点。
亿速云「云服务器」,即开即用、新一代英特尔至强铂金CPU、三副本存储NVMe SSD云盘,价格低至29元/月。点击查看>>
免责声明:本站发布的内容(图片、视频和文字)以原创、转载和分享为主,文章观点不代表本网站立场,如果涉及侵权请联系站长邮箱:is@yisu.com进行举报,并提供相关证据,一经查实,将立刻删除涉嫌侵权内容。
原文链接:https://my.oschina.net/u/4592498/blog/4451144